Modelling of intra-cellular cell decision mechanisms
Published:
I consider models of intra-cellular signalling in cell polarization, that control how a cell determines its front and back. The objective is to uncover how cell-cell interactions, intracellular signalling, forces, and adhesion affect cell polarity and lead to the emergence of collective cell behaviour, in small to large cell groups.
My hybrid PDE-cell-based-models, track 1D cells and their concentrations of intra-cellular proteins (using reaction-diffusion equations (RDEs)). The proteins Rac and Rho are known to regulate cell expansion and contraction. In my model, their concentrations determine the forces on the cell edges. Crucially my model accounts for single-cell experiment results.
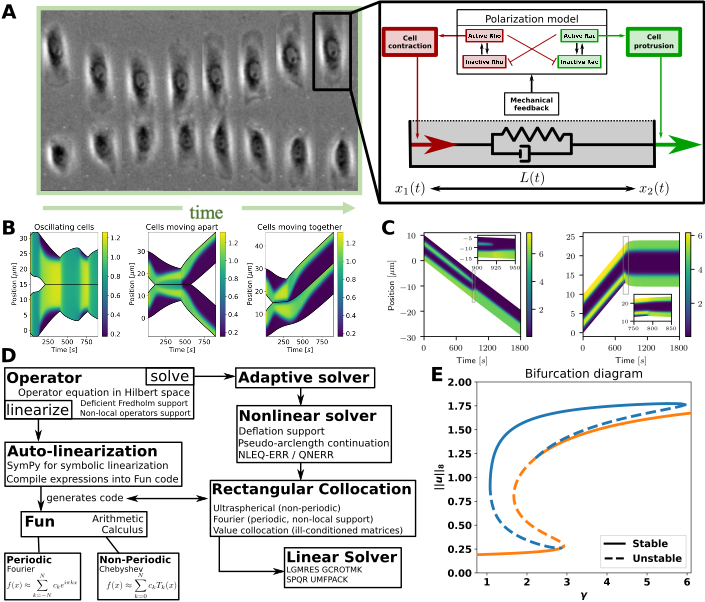
My cell repolarization / signalling model. A: Left: Cell collision experiment (Desai et al. 2013). Right: My mechanochemical model of a 1D cell has two modules: a polarization model, and visco-elastic “spring” connecting the cell’s edges. The polarization model describes intra-cellular proteins resulting in forces on the cell’s edges. B: Cell collisions in my model. C: Intra-cellular pattern formation determines cell behaviour. D: My modular ODE-iPDE analysis toolbox (based on NumPy). Symbolic linearization allows adoption to other spatial systems. E: Bifurcation diagram of a polarization model created by my toolbox. The “S-branch” consists of constant solutions, while the others are polarized states (see C).
