I am an applied mathematician, building bridges between the mathematical, computational, and biological sciences. My strong foundations in each allow me to identify key biological problems, draw on and contribute to theoretical mathematical foundations, and develop advanced computational tools.
Swarms, flocks, and human societies all exhibit complex collective behaviours. I am interested in collective cell behaviours, which I view as swarms with a twist:
- Cells are not simply point-like particles but have spatial extent;
- Interactions between cells go beyond simple attraction-repulsion; and
- Cells “live” in a regime where friction dominates over inertia.
These principles guide my work on wound healing, embryogenesis, immune response, and cancer metastasis. I use mathematical modelling and computational biology to uncover the universal principles how biological, physical, and chemical factors shape biological tissues.
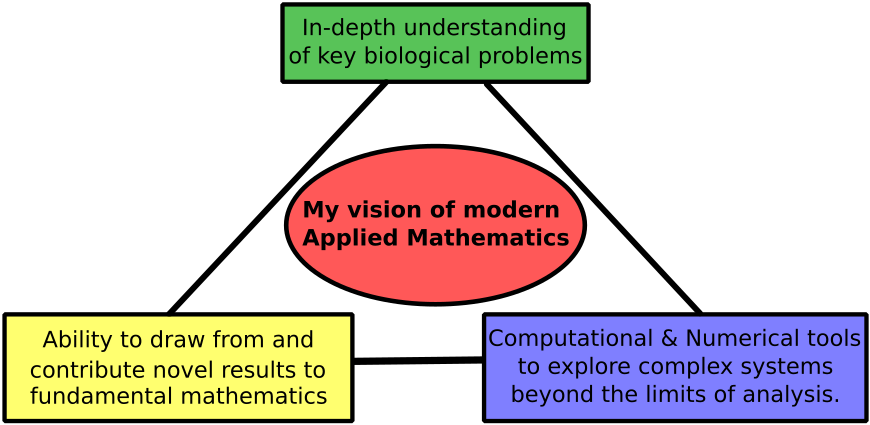
Interactions in tissues range over several orders of magnitude in time and spatial scales. Distinct mathematical frameworks are appropriate for specific levels of detail. For instance, differential equations (DEs), tracking changes in cell (or protein) densities, are suitable for describing large populations. I use dynamical systems, bifurcation theory, and group theory to analyse nonlocal DEs. On the other hand, to track the motion, behaviour, or forces produced by individual cells, a more detailed cell-based computational framework is needed. I refer to such models as cell-based models (CBMs).
Latest News
 December 2025SPECIAL ISSUE: Mathematical Modeling of Cell Motility Across Scales
December 2025SPECIAL ISSUE: Mathematical Modeling of Cell Motility Across ScalesNow open for submissions to the Journal of Mathematical Biology. Guest editors: Andreas Buttenschön, Calina Copos, and Dietmar Oelz.
 December 2025ACCEPTED: Graph-Based Preconditioners for Agent-Based Models
December 2025ACCEPTED: Graph-Based Preconditioners for Agent-Based ModelsTogether with REU student Justin Steinman, our paper on support graph preconditioners for cell-based models was accepted in the SIAM Journal on Numerical Analysis.
 October 2025PREPRINT: Competing forces of polarization and confinement generate cellular chirality in a minimal model
October 2025PREPRINT: Competing forces of polarization and confinement generate cellular chirality in a minimal modelNew preprint on chirality in cell doublets
 January 2025Teaching MATH 456: Mathematical Modeling in Fall 2025
January 2025Teaching MATH 456: Mathematical Modeling in Fall 2025I'm excited to announce I'll be teaching MATH 456: Mathematical Modeling in Fall 2025, focusing on mathematical approaches to modeling cellular phenomena.
 October 2024PREPRINT: Graph-Based Preconditioners for Agent-Based Models
October 2024PREPRINT: Graph-Based Preconditioners for Agent-Based ModelsTogether with REU student Justin Steinman, we published our first lab pre-print on support graph preconditioners for cell-based models.
 October 2024ACCEPTED: The Cell Cluster Instability
October 2024ACCEPTED: The Cell Cluster InstabilityHow cells stay together; a mechanism for maintenance of a robust cluster explored by local and nonlocal continuum models.
 June 2024PREPRINT: The Cell Cluster Instability
June 2024PREPRINT: The Cell Cluster InstabilityHow cells stay together; a mechanism for maintenance of a robust cluster explored by local and nonlocal continuum models.
 January 2021BOOK: on Bifurcations and Symmetries Published
January 2021BOOK: on Bifurcations and Symmetries PublishedOur book on bifurcations and symmetries in non-local cell adhesion models is now available from Springer.
 January 2020PAPER: Review on Cell Migration Models Published
January 2020PAPER: Review on Cell Migration Models PublishedOur comprehensive review on computational and mathematical models of cell migration is now published in PLOS Computational Biology.
