# Math 551: uMass Amherst Fall 2022
import numpy as np
import scipy as sp
import scipy.linalg as LA
import math
import matplotlib.pyplot as plt
from prettytable import PrettyTable, MARKDOWNPoisson’s Equations¶
The test program in this notebook is the Poisson equation
with Dirichlet boundary conditions.
import scipy.sparse as sps
# Disc points
L = 1
N = 4
h = 1./(N-1)
# Create differentiation matrices
D2 = sps.diags([-1, 2, -1], [-1, 0, 1], shape=(N, N))
# Now assemble 2D matrices
Ix = sps.eye(N)
Iy = sps.eye(N)
D2_x = sps.kron(Ix, D2)
D2_y = sps.kron(D2, Iy)
D2 = D2_x + D2_y
print(D2.shape)(16, 16)
# Implement a Poisson solver
# 1. Implement a LinearOperator that represents the finite difference matrix.
from scipy.sparse.linalg import LinearOperator
class Laplacian(LinearOperator):
""" Poisson operator """
def __init__(self, N, dtype=None, *args, **kwargs):
""" N is the grid size """
L = kwargs.pop('L', 1.)
self.h = L / (N - 1)
# Create differentiation matrices
D2 = sps.diags([-1, 2, -1], [-1, 0, 1], shape=(N, N))
# Now assemble 2D matrices
Ix = sps.eye(N)
Iy = sps.eye(N)
D2_x = sps.kron(Ix, D2)
D2_y = sps.kron(D2, Iy)
self.D2 = D2_x + D2_y
# Call constructor of base
super().__init__(dtype=np.dtype(dtype), shape=(N**2, N**2))
def _matvec(self, x):
""" Computes Laplacian using central finite difference scheme """
return self.D2 @ xdef create_problem(N, L=2.*np.pi):
def g(x, y):
return np.cos(2. * x) + np.cos(2. * y) + np.cos(x) * np.cos(y)
# Now we have Dirichlet BCs -> only need to solve on the internal grid.
# uniform points in both directions
x = np.linspace(0, L, N)
xx, yy = np.meshgrid(x[1:-1], x[1:-1])
# Create right hand vector
gij = g(xx, yy).flatten()
# Create Laplacian
LN = Laplacian(N-2, L=L)
h = L / (N - 3)
# Return the system
return LN, h**2 * gij
def plot_soln(xs, L=2.*np.pi):
N2 = int(np.sqrt(xs.shape[0]))
N = N2 + 2
# Plot the solution
x = np.linspace(0, L, N)
xx, yy = np.meshgrid(x, x)
zz = np.zeros_like(xx)
zz[1:-1, 1:-1] = xs.reshape((N-2, N-2))
plt.pcolormesh(xx, yy, zz)
plt.xlim([0, L])
plt.ylim([0, L])
plt.xlabel('$x$')
plt.ylabel('$y$')
plt.colorbar()Jacobi Method¶
b1 = 1.0
b2 = 1.0
xr = np.ones(2)
def Jacobi(x):
x1, x2 = x
return np.array([
0.5 * (x2 + 1),
0.5 * (x1 + 1)
])
x = np.zeros(2)
for i in range(5):
x = Jacobi(x)
print('x[{0:d}] = {1:.4g}, {2:.4g}, {3:.4g}'.format(i, x[0], x[1], np.max(np.abs(x - xr))))x[0] = 0.5, 0.5, 0.5
x[1] = 0.75, 0.75, 0.25
x[2] = 0.875, 0.875, 0.125
x[3] = 0.9375, 0.9375, 0.0625
x[4] = 0.9688, 0.9688, 0.03125
b1 = 3.0
b2 = -3.0
xr = np.ones(2)
xr[1] = -1
def Jacobi(x):
x1, x2 = x
return np.array([
0.25 * (3 - x2),
-0.25 * (x1 + 3)
])
x = np.zeros(2)
for i in range(5):
x = Jacobi(x)
print('x[{0:d}] = {1:.4g}, {2:.4g}, {3:.4g}'.format(i, x[0], x[1], np.max(np.abs(x - xr))))x[0] = 0.75, -0.75, 0.25
x[1] = 0.9375, -0.9375, 0.0625
x[2] = 0.9844, -0.9844, 0.01562
x[3] = 0.9961, -0.9961, 0.003906
x[4] = 0.999, -0.999, 0.0009766
Gauss-Seidel¶
b1 = 1.0
b2 = 1.0
xr = np.ones(2)
def GS(x):
x1, x2 = x
xk1 = 0.5 * (x2 + 1)
return np.array([
xk1,
0.5 * (xk1 + 1.0)
])
x = np.zeros(2)
for i in range(5):
x = GS(x)
print('x[{0:d}] = {1:.4g}, {2:.4g}, {3:.4g}'.format(i, x[0], x[1], np.max(np.abs(x - xr))))x[0] = 0.5, 0.75, 0.5
x[1] = 0.875, 0.9375, 0.125
x[2] = 0.9688, 0.9844, 0.03125
x[3] = 0.9922, 0.9961, 0.007812
x[4] = 0.998, 0.999, 0.001953
b1 = 3.0
b2 = -3.0
xr = np.ones(2)
xr[1] = -1
def GS(x):
x1, x2 = x
return np.array([
0.25 * (3 - x2),
(x2 - 15)/16.
])
x = np.zeros(2)
for i in range(5):
x = GS(x)
print('x[{0:d}] = {1:.4g}, {2:.4g}, {3:.4g}'.format(i, x[0], x[1], np.max(np.abs(x - xr))))x[0] = 0.75, -0.9375, 0.25
x[1] = 0.9844, -0.9961, 0.01562
x[2] = 0.999, -0.9998, 0.0009766
x[3] = 0.9999, -1, 6.104e-05
x[4] = 1, -1, 3.815e-06
SOR¶
b1 = 1.0
b2 = 1.0
xr = np.ones(2)
# SOR parameter omega in (1, 2)
omega = 1.5
def SOR(x):
x1, x2 = x
# Do the GS step
xk1 = 0.5 * (x2 + 1.)
xk2 = 0.5 * (xk1 + 1.)
# Then do SOR
xk1_SOR = (1. - omega) * x1 + omega * xk1
xk2_SOR = (1. - omega) * x2 + omega * xk2
return np.array([xk1_SOR, xk2_SOR])
x = np.zeros(2)
for i in range(5):
x = SOR(x)
print('x[{0:d}] = {1:.4g}, {2:.4g}, {3:.4g}'.format(i, x[0], x[1], np.max(np.abs(x - xr))))x[0] = 0.75, 1.125, 0.25
x[1] = 1.219, 0.9844, 0.2188
x[2] = 0.8789, 1.002, 0.1211
x[3] = 1.062, 0.9998, 0.06201
x[4] = 0.9688, 1, 0.03119
Gradient Descent¶
import scipy.sparse as sps
from scipy.sparse.linalg import LinearOperator
#from pylops import LinearOperator
class MatrixVectorProduct(LinearOperator):
def __init__(self, A, dtype=None):
self.A = A
super().__init__(dtype=np.dtype(dtype), shape=A.shape)
def _matvec(self, x):
""" Implements the matrix vector product """
return self.A @ x
# Example
A = np.array([[7, 3, 1], [3, 10, 2], [1, 2, 15]])
b = np.array([28, 31, 22])
# Solution is [3, 2, 1]
# Create the linear operator
Ax = MatrixVectorProduct(A)import numpy as np
import matplotlib.pyplot as plt
A = np.array([[3, 2], [2, 6]])
b = np.array([2, -8])
c = 0
x = np.array([2, -2])
print('Ax = ', A @ x)
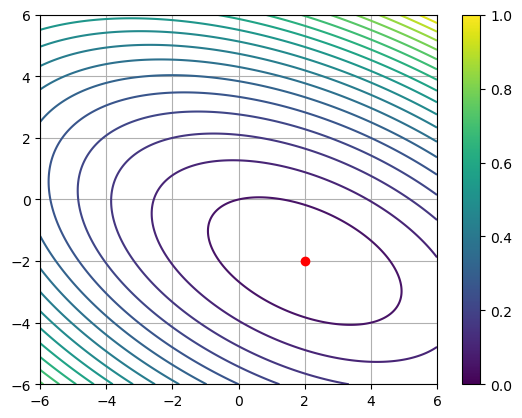
def f(x, y):
return 0.5 * (3. * x**2 + 4. * x * y + 6 * y**2) - 2 * x + 8 * y
xs = np.linspace(-6, 6, 1000)
xx, yy = np.meshgrid(xs, xs)
plt.contour(xx, yy, f(xx, yy), 25)
plt.scatter(2, -2, color='r', zorder=5)
plt.grid()
plt.colorbar()Ax = [ 2 -8]
def gdesc(A, b, tol=1e-12, norm=lambda x: LA.norm(x, ord=2), *args, **kwargs):
# make sure types are right
A = sps.linalg.aslinearoperator(A)
b = b.astype(float)
# In this case we know what r will be!
xk = kwargs.pop('x0', np.zeros_like(b))
rk = b - A.matvec(xk)
dk = np.dot(rk, rk)
bd = np.dot(b, b)
k = 0
pts = [xk]
while dk > tol*tol*bd:
sk = A.matvec(rk)
ak = dk / np.dot(rk, sk)
xk = xk + ak * rk
pts.append(xk)
rk = rk - ak * sk
dk = np.dot(rk, rk)
k += 1
return xk, norm(rk), k, pts
# Example
# A = np.array([[7, 3, 1], [3, 10, 2], [1, 2, 15]])
# b = np.array([28, 31, 22])
# Solution is [3, 2, 1]
# Create the linear operator
Ax = MatrixVectorProduct(A)
# Test the method
x, r, k, pts = gdesc(Ax, b)
print('||B - Ax|| = ', np.max(np.abs(A @ x - b)))
print('x = ', x)
print('r = ', r)
print('k = ', k)||B - Ax|| = 5.019540338935258e-12
x = [ 2. -2.]
r = 5.1741851798292685e-12
k = 47
pts = np.asarray(pts)
xs = np.linspace(-2, 6, 1000)
ys = np.linspace(-4, 1, 100)
xx, yy = np.meshgrid(xs, ys)
plt.contour(xx, yy, f(xx, yy), 25)
plt.plot(pts[:, 0], pts[:, 1], color='k')
plt.grid()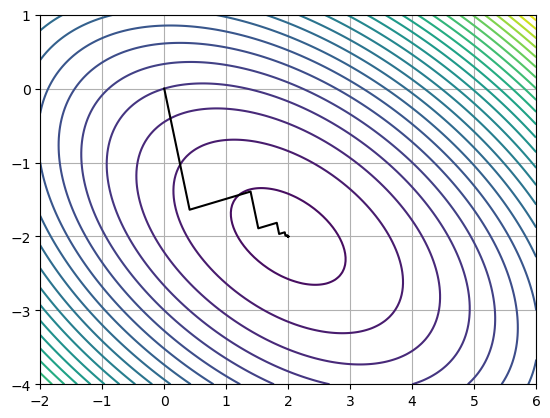
Conjugate gradient¶
def cg(A, b, tol=1e-12, norm=lambda x: LA.norm(x, ord=2), *args, **kwargs):
# make sure types are right
A = sps.linalg.aslinearoperator(A)
b = b.astype(float)
# In this case we know what r will be!
xk = kwargs.pop('x0', np.zeros_like(b))
rk = b - A.matvec(xk)
dk = np.dot(rk, rk)
bd = np.dot(b, b)
pk = rk.copy()
pts = [xk]
ps = [pk]
k = 0
while dk > tol*tol*bd:
sk = A.matvec(pk)
ak = dk / np.dot(pk, sk)
xk = xk + ak * pk
pts.append(xk)
rk = rk - ak * sk
dk1 = np.dot(rk, rk)
pk = rk + (dk1 / dk) * pk
ps.append(pk)
dk = dk1
k += 1
return xk, norm(rk), k, pts, ps
# Example
# A = np.array([[7, 3, 1], [3, 10, 2], [1, 2, 15]])
# b = np.array([28, 31, 22])
# Solution is [3, 2, 1]
# Create the linear operator
Ax = MatrixVectorProduct(A)
# Test the method
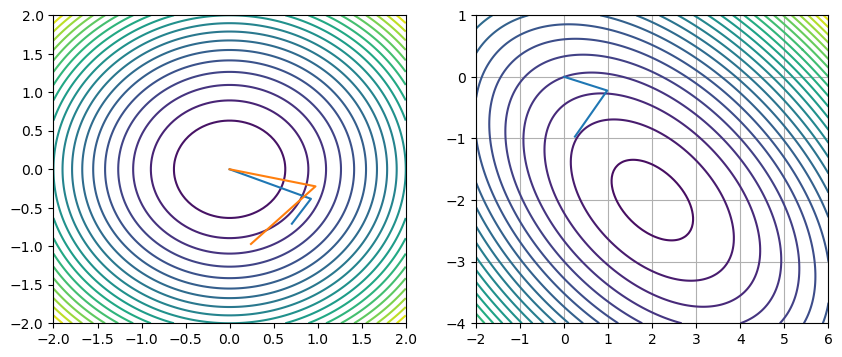
x, r, k, pts, ps = cg(Ax, b)
print('||B - Ax|| = ', np.max(np.abs(A @ x - b)))
print('x = ', x)
print('r = ', r)
print('k = ', k)||B - Ax|| = 0.0
x = [ 2. -2.]
r = 9.930136612989092e-16
k = 2
A1 = (1./14) * np.array([[6, -2], [-2, 3]])
pts = np.asarray(pts)
ps = np.asarray(ps)
qs = np.zeros_like(ps)
for i in range(ps.shape[1]):
ps[i, :] /= np.linalg.norm(ps[i, :], ord=2)
qs[i, :] = A1 @ ps[i, :]
qs[i, :] /= np.linalg.norm(qs[i, :], ord=2)
fig, axs = plt.subplots(1, 2, figsize=(10, 4))
xs = np.linspace(-2, 2, 100)
ys = np.linspace(-2, 2, 100)
xx, yy = np.meshgrid(xs, ys)
axs[0].contour(xx, yy, xx**2 + yy**2, 25)
axs[0].plot(qs[:, 0], qs[:, 1])
axs[0].plot(ps[:, 0], ps[:, 1])
xs = np.linspace(-2, 6, 1000)
ys = np.linspace(-4, 1, 100)
xx, yy = np.meshgrid(xs, ys)
axs[1].contour(xx, yy, f(xx, yy), 25)
axs[1].plot(ps[:, 0], ps[:, 1])
print(ps)
plt.grid()[[ 2.42535625e-01 -9.70142500e-01]
[ 9.75132856e-01 -2.21621104e-01]
[-8.88178420e-16 -4.44089210e-16]]
pts = np.asarray(pts)
xs = np.linspace(-2, 6, 1000)
ys = np.linspace(-4, 1, 100)
xx, yy = np.meshgrid(xs, ys)
plt.contour(xx, yy, f(xx, yy), 25)
# Plot path
plt.plot(pts[:, 0], pts[:, 1], color='k')
plt.grid()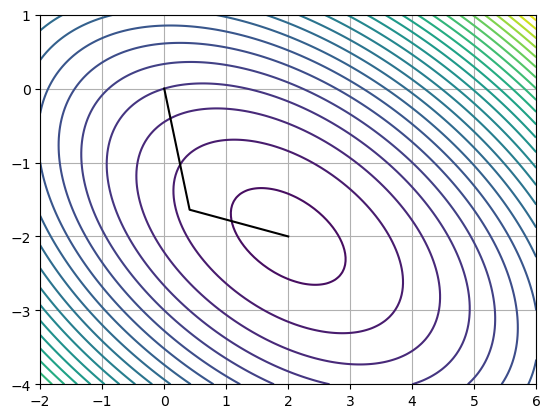
Gradient Descent 2D Poisson¶
L, b = create_problem(N=63)
# Solve the linear system
xs, r, k, _ = gdesc(L, b, tol=1e-12)
print('Linear system solved in {0:d} iterations with ||r|| = {1:.5g}.'.format(k, r))
plot_soln(xs)Linear system solved in 18566 iterations with ||r|| = 7.3953e-13.
Conjugate gradient and Poisson¶
L, b = create_problem(N=63)
print(L.shape)
# Solve the linear system
xs, r, k, _, _ = cg(L, b, tol=1e-12)
print('Linear system solved in {0:d} iterations with ||r|| = {1:.5g}.'.format(k, r))
plot_soln(xs)(3721, 3721)
Linear system solved in 140 iterations with ||r|| = 4.9752e-13.

Preconditioned conjugate gradient¶
def pcg(A, P, b, tol=1e-12, norm=lambda x: LA.norm(x, ord=2), *args, **kwargs):
# make sure types are right
A = sps.linalg.aslinearoperator(A)
b = b.astype(float)
# In this case we know what r will be!
xk = kwargs.pop('x0', np.zeros_like(b))
rk = b - A.matvec(xk)
hk = P.solve(rk)
dk = np.dot(rk, hk)
bd = np.dot(b, P.solve(b))
pk = hk.copy()
k = 0
while dk > tol*tol*bd:
sk = A.matvec(pk)
ak = dk / np.dot(pk, sk)
xk = xk + ak * pk
rk = rk - ak * sk
hk = P.solve(rk)
dk1 = np.dot(rk, hk)
pk = hk + (dk1 / dk) * pk
dk = dk1
k += 1
return xk, norm(rk), k
# Example
A = np.array([[7, 3, 1], [3, 10, 2], [1, 2, 15]])
b = np.array([28, 31, 22])
# Solution is [3, 2, 1]
# Create the linear operator
Ax = MatrixVectorProduct(A)
# Create the preconditioner by defauly uses ILU -> TODO symmetrify i.e. Cholesky
P = sps.linalg.spilu(A, drop_tol=1e-2)
# Test the method
x, r, k = pcg(Ax, P, b)
print('||B - Ax|| = ', np.max(np.abs(A @ x - b)))
print('x = ', x)
print('r = ', r)
print('k = ', k)||B - Ax|| = 7.105427357601002e-15
x = [3. 2. 1.]
r = 8.702335715267317e-15
k = 1
/usr/lib/python3.10/site-packages/scipy/sparse/linalg/_dsolve/linsolve.py:437: SparseEfficiencyWarning: spilu converted its input to CSC format
warn('spilu converted its input to CSC format',
L, b = create_problem(N=63)
# Assemb le a precond
P = sps.linalg.spilu(L.D2.tocsc(), drop_tol=0.01)
print('Solve!')
# Solve the linear system
xs, r, k = pcg(L, P, b, tol=1e-12)
print('Linear system solved in {0:d} iterations with ||r|| = {1:.5g}.'.format(k, r))
plot_soln(xs)Solve!
Linear system solved in 365 iterations with ||r|| = 4.0499e-12.

Multigrid preconditioned conjugate gradient¶
http://
Let’s learn something about multigrid methods. The equation at grid level is given by
We also need to transfer maps: (1) The restriction map which maps a vector from a finer grid to a coarsers grid denoted by , and (2) the prolongation operator which maps coarser grids to finer grids, denoted by .
The restriction operator is defined as the adjoint of prolongation, that is, it satisfies
where is a scalar constant.
Two-grid methods¶
A linear equation is considered. Let be the relaxant matrix and is an approximate vector.
Relaxation
Coarse-grid correction
Map back to fine grid
Post-smoothing step
Let’s split the matrix as follows
when using Jacobi P will contain the diagonal eleemnts. Then update the vector
We do iterations of this. Then the matrix representing these iterations is given by .
# Poisson equation
N = 4
D1 = sps.diags([-1, 2, -1], [-1, 0, 1], shape=(N, N))
print(type(D1))
print(D1.todense())
D2_x = sps.diags([-1, 2, -1], [-1, 0, 1], shape=(N, N))
D2_y = sps.diags([-1, 2, -1], [-1, 0, 1], shape=(N, N))
D2 = sps.kron(sps.eye(N), D2_x) + sps.kron(D2_y, sps.eye(N))
D2 = D2.todia()# Inter grid operations in 1D
# Define prolongation operation
# I_H^h : Omega_H -> Omega_h
# [x_0, x_1, ..., x_{N+1}]
# Number of internal points is odd -> N-1 odd -> N+1 odd -> N even
N = 5
# v^h = I_2h^h v^{2h}
# v^h_2j = 2_j^2h
# v^h_2j+1 = (v^2h_j + v^2h_j+1) / 2
I = sps.lil_array((N+2, (N+1)//2))
cc = np.array([1, 2, 1])
for k in range((N+1)//2):
kk = 2*k
I[kk:kk+3, k] = cc
# Scale
I = I[1:-1, :]
I *= 0.5
# Cmpute inverse
R = I.transpose()
R *= 0.5
R[0, 1] = 0.
R[-1, -2 ] = 0.
R[0, 0] *= 2.
R[-1, -1] *= 2
print('I = ')
print(I.todense())
print()
print('R = ')
print(R.todense())
xs = np.linspace(0, 1, (N+1)//2)
plt.scatter(xs, np.zeros_like(xs), color='r')
ys = I @ xs
zs = R @ ys
print(xs)
print(ys)
print(zs)
plt.scatter(ys, .1 *np.ones_like(ys), color='b')
plt.scatter(zs, .2 *np.ones_like(zs), color='k')
plt.ylim([-0.1, .3])def smooth(steps, A, x0, f):
# TODO implement damped Jacobi / GS.
for i in range(steps):
x0 += B @ (f - A @ x0)
return x0# 2D interpolation matrix is the tensor product of the interpolator
# in both directions
I2D = sps.kron(I, I)
# Test set
x = np.linspace(0, 1, 3)
y = np.linspace(0, 1, 3)
xx, yy = np.meshgrid(x, y)
print(I2D.shape)
print(xx.shape)
A = np.arange(9).reshape((3, 3))
plt.pcolormesh(xx, yy, A)
plt.colorbar()
# Test set
x = np.linspace(0, 1, 5)
y = np.linspace(0, 1, 5)
xx, yy = np.meshgrid(x, y)
B = I2D @ A.flatten()
plt.pcolormesh(xx, yy, B.reshape((5, 5)))
plt.colorbar()# CF based AMG -> get strength matrix
def CfStrength(A):
n, m = A.shape
assert n == m, ''
from sympy import pycode, tan, Symbol, srepr, N, sympify, lambdify
x = Symbol('x')
expr_str = 'tan(x) + 1'
expr1 = sympify(expr_str)
# lam_str = lambdastr(x, expr1)
print(lam_str)
lam = lambdify(x, expr1)
print(lam(1))
py_str = pycode(expr1)
print('Python code = ')
print(py_str)
expr = tan(x) + 1
# Slow symp eval
res = expr.subs(x, 2)
print(res)
print(N(res))
srepr(expr)